diff --git a/.github/workflows/test-coverage.yaml b/.github/workflows/test-coverage.yaml
index f0923535..44b98225 100644
--- a/.github/workflows/test-coverage.yaml
+++ b/.github/workflows/test-coverage.yaml
@@ -1,11 +1,11 @@
name: Test Coverage
on:
- pull_request:
- push:
- branches:
- - main
- workflow_dispatch:
+ pull_request:
+ push:
+ branches:
+ - main
+ workflow_dispatch:
jobs:
test:
@@ -40,7 +40,9 @@ jobs:
env:
PTO_CI_MODE: 1
run: |
- pytest -v --cov=pytissueoptics --cov-report=xml --cov-report=html
+ pytest --cov --cov-branch --cov-report=term --cov-report=xml
- name: Upload coverage report
- uses: codecov/codecov-action@v4
+ uses: codecov/codecov-action@v5
+ with:
+ token: ${{ secrets.CODECOV_TOKEN }}
diff --git a/.github/workflows/tests.yaml b/.github/workflows/tests.yaml
index f29adb6f..2bac20ea 100644
--- a/.github/workflows/tests.yaml
+++ b/.github/workflows/tests.yaml
@@ -5,6 +5,8 @@ on:
push:
branches:
- main
+ schedule:
+ - cron: "0 20 * * 4" # Every Thursday @ 20:00 UTC (15:00 EST)
workflow_dispatch:
jobs:
diff --git a/README.md b/README.md
index 44c9611b..38b0ed7f 100644
--- a/README.md
+++ b/README.md
@@ -1,45 +1,31 @@
-
PyTissueOptics
+A hardware-accelerated Python module to simulate light transport in arbitrarily complex 3D media with ease.
+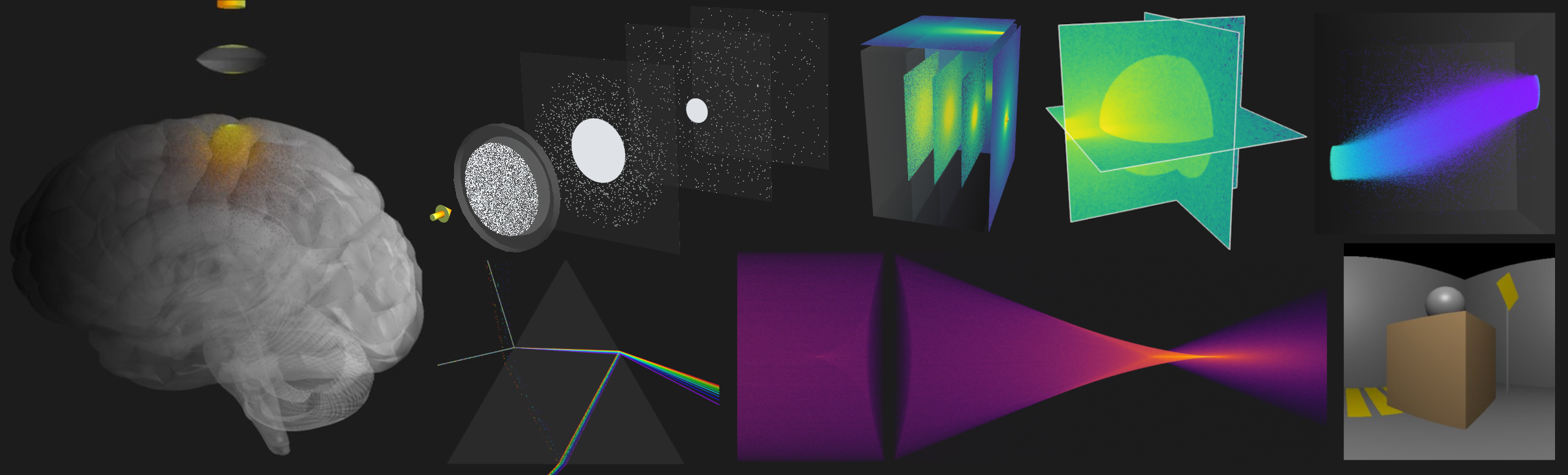 -
-Monte Carlo simulations of light transport made easy.
-
- -
-
+[](https://github.com/DCC-Lab/pytissueoptics/actions/workflows/tests.yaml) [](https://codecov.io/gh/DCC-Lab/pytissueoptics) [](https://www.codefactor.io/repository/github/DCC-Lab/pytissueoptics) [](LICENSE)
-This python package is an object-oriented implementation of Monte Carlo modeling for light transport in diffuse media.
-The package is very **easy to set up and use**, and its mesh-based approach makes it a **polyvalent** tool to simulate light transport in arbitrarily complex scenes.
-The package offers both a native Python implementation and a hardware-accelerated version using OpenCL.
+This python package is a fast and flexible implementation of Monte Carlo modeling for light transport in diffuse media.
+The package is **easy to set up and use**, and its mesh-based approach makes it a polyvalent tool to simulate
+light transport in **arbitrarily complex scenes**. The package offers both a native Python implementation
+and a **hardware-accelerated** version using OpenCL which supports most GPUs and CPUs.
-As discussed in the [why use this package](#why-use-this-package) section, computation time isn't the only variable at play. This code is **easy to understand**, **easily scalable** and **very simple to modify** for your need. It was designed with **research and education** in mind.
+Designed with **research and education** in mind, the code aims to be clear, modular, and easy to extend for a wide range of applications.
## Notable features
-- Arbitrarily complex 3D environments.
-- Import external 3D models (.OBJ).
-- Great data visualization with `Mayavi`.
-- Multi-layered tissues.
-- Hardware acceleration with `OpenCL`.
-- Accurate Fresnel reflection and refraction with surface smoothing.
-- Discard 3D data (auto-binning to 2D views).
-- Independent 3D graphics framework under `scene`.
-
-## Table of Contents
-- [Installation](#installation)
-- [Getting Started](#getting-started)
-- [Hardware Acceleration](#hardware-acceleration)
-- [Why Use This Package](#why-use-this-package)
-- [Examples](#examples)
+- Supports arbitrarily complex **mesh-based** 3D environments.
+- **Normal smoothing** for accurate modeling of curved surfaces like lenses.
+- Per-photon data points of deposited energy, fluence rate and intersection events.
+- **Hardware accelerated** with `OpenCL` using [PyOpenCL](https://github.com/inducer/pyopencl).
+- Photon traces & detectors.
+- Import **external 3D models** (`.OBJ`).
+- Many 3D visualization options built with [Mayavi](https://github.com/enthought/mayavi).
+- Low memory mode with auto-binning to 2D views.
+- **Reusable graphics framework** to kickstart other raytracing projects like [SensorSim](https://github.com/JLBegin/SensorSim).
## Installation
-Requires Python >=3.9 installed on the device.
-
-### Installing the latest release
-> Currently, this `pip` version is outdated. We recommend installing the development version.
-```shell
-python -m pip install --upgrade pip
-python -m pip install --upgrade pytissueoptics
-```
+Requires Python 3.9+ installed.
-### Installing the development version
+> Currently, the `pip` version is outdated. We recommend installing the development version.
1. Clone the repository.
2. Create a virtual environment inside the repository with `python -m venv venv`.
3. Activate the virtual environment.
@@ -49,80 +35,30 @@ python -m pip install --upgrade pytissueoptics
5. Install the package requirements with `python -m pip install -e .[dev]`.
## Getting started
-A command-line interface is available to help you run examples and tests.
-```shell
-python -m pytissueoptics --help
-python -m pytissueoptics --list
-python -m pytissueoptics --examples 1,2,3
-python -m pytissueoptics --tests
-```
-To launch a simple simulation on your own, follow these steps.
-1. Import the `pytissueoptics` module
-2. Define the following objects:
- - `scene`: a `ScatteringScene` object, which defines the scene and the optical properties of the media, or use a pre-defined scene from the `samples` module. The scene takes in a list of `Solid` as its argument. These `Solid` will have a `ScatteringMaterial` and a position. This is clear in the examples below.
- - `source`: a `Source` object, which defines the source of photons.
- - `logger`: an `EnergyLogger` object, which logs the simulation progress ('keep3D=False' can be set to auto-bin to 2D views).
-3. Propagate the photons in your `scene` with `source.propagate`.
-4. Define a `Viewer` object and display the results by calling the desired methods. It offers various visualizations of the experiment as well as a statistics report.
-
-Here's what it might look like:
-```python
-from pytissueoptics import *
-
-material = ScatteringMaterial(mu_s=3.0, mu_a=1.0, g=0.8, n=1.5)
-
-tissue = Cuboid(a=1, b=3, c=1, position=Vector(2, 0, 0), material=material)
-scene = ScatteringScene([tissue])
-
-logger = EnergyLogger(scene)
-source = PencilPointSource(position=Vector(-3, 0, 0), direction=Vector(1, 0, 0), N=1000)
-source.propagate(scene, logger)
+A **command-line interface** is available to quicky run a simulation from our pool of examples:
-viewer = Viewer(scene, source, logger)
-viewer.reportStats()
-viewer.show3D()
```
-Check out the `pytissueoptics/examples` folder for more examples on how to use the package.
-
-## Hardware acceleration
-```python
-source = DivergentSource(useHardwareAcceleration=True, ...)
+python -m pytissueoptics --help
```
-Hardware acceleration can offer a speed increase factor around 1000x depending on the scene.
-By default, the program will try to use hardware acceleration if possible, which will require OpenCL drivers for your hardware of choice.
-NVIDIA and AMD GPU drivers should contain their corresponding OpenCL driver by default.
-To force the use of the native Python implementation, set `useHardwareAcceleration=False` when creating a light `Source`.
-
-Follow the instructions on screen to get setup properly for the first hardware accelerated execution which will offer
-to run a benchmark test to determine the ideal number of work units for your hardware.
-
-## Why use this package
-It is known, as April 2022, Python is **the most used** language ([Tiobe index](https://www.tiobe.com/tiobe-index/)).
-This is due to the ease of use, the gentle learning curve, and growing community and tools. There was a need for
-such a package in Python, so that not only long hardened C/C++ programmers could use the power of Monte Carlo simulations.
-It is fairly reasonable to imagine you could start a calculation in Python in a few minutes, run it overnight and get
-an answer the next day after a few hours of calculations. It is also reasonable to think you could **modify** the code
-yourself to suit your exact needs! (Do not attempt this in C). This is the solution that the CPU-based portion of this package
-offers you. With the new OpenCL implementation, speed is not an issue anymore, so using `pytissueoptics` should not even be a question.
-
-### Known limitations
-1. It uses Henyey-Greenstein approximation for scattering direction because it is sufficient most of the time.
-2. Reflections are specular, which does not accounts for the roughness of materials. It is planned to implement Bling-Phong reflection model in a future release.
-
-## Examples
-### Multi-layered phantom tissue
-Located at `pytissueoptics/examples/rayscattering/ex01.py`.
-Using a pre-defined tissue from the `samples` module.
+You can kick start your first simulation using one of our **pre-defined scene** under the `samples` module.
```python
-N = 500000
+from pytissueoptics import *
+
+# Define (scene, source, logger)
+N = 500_000
scene = samples.PhantomTissue()
-source = DivergentSource(position=Vector(0, 0, -0.1), direction=Vector(0, 0, 1), N=N, diameter=0.2, divergence=np.pi / 4)
+source = DivergentSource(
+ position=Vector(0, 0, -0.1), direction=Vector(0, 0, 1), N=N, diameter=0.2, divergence=0.78
+)
logger = EnergyLogger(scene)
+
+# Run
source.propagate(scene, logger=logger)
+# Stats & Visualizations
viewer = Viewer(scene, source, logger)
viewer.reportStats()
@@ -132,56 +68,73 @@ viewer.show2D(View2DSurfaceZ(solidLabel="middleLayer", surfaceLabel="interface0"
viewer.show1D(Direction.Z_POS)
viewer.show3D()
```
+#### Expected output
+```
+Report of solid 'backLayer'
+ Absorbance: 67.78% (10.53% of total power)
+ Transmittance at 'backLayer_back': 22.4%
+ Transmittance at 'interface0': 4.9%
+ ...
+```
+
+
-#### Default figures generated

-#### Discarding the 3D data
-When the raw simulation data gets too large, the 3D data can be automatically binned to pre-defined 2D views.
+#### Scene definition
+Here is the explicit definition of the above scene sample. We recommend you look at other examples to get familiar with the API.
```python
-logger = EnergyLogger(scene, keep3D=False)
-```
-All 2D views are unchanged, because they are included in the default 2D views tracked by the EnergyLogger.
-The 1D profile and stats report are also properly computed from the stored 2D data.
-The 3D display will auto-switch to Visibility.DEFAULT_2D which includes Visibility.VIEWS with ViewGroup.SCENE (XYZ projections of the whole scene) visible by default.
-
-
-#### Display some 2D views with the 3D point cloud
-The argument `viewsVisibility` can accept a `ViewGroup` tag like SCENE, SURFACES, etc., but also a list of indices for fine control. You can list all stored views with `logger.listViews()` or `viewer.listViews()`.
-Here we toggle the visibility of 2D views along the default 3D visibility (which includes the point cloud).
-```python
-logger = EnergyLogger(scene)
-[...]
-viewer.show3D(visibility=Visibility.DEFAULT_3D | Visibility.VIEWS, viewsVisibility=[0, 1])
+materials = [
+ ScatteringMaterial(mu_s=2, mu_a=1, g=0.8, n=1.4),
+ ScatteringMaterial(mu_s=3, mu_a=1, g=0.8, n=1.7),
+ ScatteringMaterial(mu_s=2, mu_a=1, g=0.8, n=1.4),
+]
+w = 3
+frontLayer = Cuboid(a=w, b=w, c=0.75, material=materials[0], label="frontLayer")
+middleLayer = Cuboid(a=w, b=w, c=0.5, material=materials[1], label="middleLayer")
+backLayer = Cuboid(a=w, b=w, c=0.75, material=materials[2], label="backLayer")
+layerStack = backLayer.stack(middleLayer, "front").stack(frontLayer, "front")
+scene = ScatteringScene([layerStack])
```
-
-#### Display custom 2D views
-If `keep3D=False`, the custom views (like slices, which are not included in the default views) have to be added to the logger before propagation like so:
-```python
-logger = EnergyLogger(scene, keep3D=False)
-logger.addView(View2DSliceZ(position=0.5, thickness=0.1, limits=((-1, 1), (-1, 1))))
-logger.addView(View2DSliceZ(position=1, thickness=0.1, limits=((-1, 1), (-1, 1))))
-logger.addView(View2DSliceZ(position=1.5, thickness=0.1, limits=((-1, 1), (-1, 1))))
-```
-If the logger keeps track of 3D data, then it's not a problem and the views can be added later with `logger.addView(customView)` or implicitly when asking for a 2D display from `viewer.show2D(customView)`.
-
+#### Hardware acceleration
-#### Display interactive 3D volume slicer
-Requires 3D data.
-```python
-viewer.show3DVolumeSlicer()
-```
-
+Depending on your platform and GPU, you might already have OpenCL drivers installed, which should work out of the box.
+Run a PyTissueOptics simulation first to see your current status.
+
+> Follow the instructions on screen to get setup properly. It will offer to run a benchmark test to determine the ideal number of work units for your hardware.
+For more help getting OpenCL to work, refer to [PyOpenCL's documentation](https://documen.tician.de/pyopencl/misc.html#enabling-access-to-cpus-and-gpus-via-py-opencl) on the matter. Note that you can disable hardware acceleration at any time with `disableOpenCL()` or by setting the environment variable `PTO_DISABLE_OPENCL=1`.
+
+## Examples
+
+All examples can be run using the CLI tool:
-#### Save and append simulation results
-The `EnergyLogger` data can be saved to file. This can also be used along with `keep3D=False` to only save 2D data. Every time the code is run, the previous data is loaded and extended. This is particularly useful to propagate a very large amount of photons (possibly infinite) in smaller batches so the hardware doesn't run out of memory.
-```python
-[...]
-logger = EnergyLogger(scene, "myExperiment.log", keep3D=False)
-source.propagate(scene, logger)
-logger.save()
```
+python -m pytissueoptics --list
+python -m pytissueoptics --examples 1,2,3
+```
+
+1. [Scene sample](/pytissueoptics/examples/rayscattering/ex01.py)
+2. [Infinite medium](/pytissueoptics/examples/rayscattering/ex02.py)
+3. [Optical lens & saving progress](/pytissueoptics/examples/rayscattering/ex03.py)
+4. [Custom layer stack](/pytissueoptics/examples/rayscattering/ex04.py)
+5. [Sphere in cube](/pytissueoptics/examples/rayscattering/ex05.py)
+6. [Sampling volume simulation](/pytissueoptics/examples/rayscattering/ex06.py)
+
+Other scene and benchmark examples are available under [/examples](/pytissueoptics/examples), including:
+- [External 3D model](/pytissueoptics/examples/scene/example3.py)
+- [Solid transforms](/pytissueoptics/examples/scene/example1.py)
+- [Lenses](/pytissueoptics/examples/scene/example4.py)
+- [Skin vessel benchmark](/pytissueoptics/examples/benchmarks/skinvessel.py)
+- [Spherical shells benchmark](/pytissueoptics/examples/benchmarks/sphshells.py)
+
+---
+
+### Known limitations
+
+1. It uses Henyey-Greenstein approximation for scattering direction because it is sufficient most of the time.
+2. Reflections are specular, which does not account for the roughness of materials. A Bling-Phong reflection model could be added in a future release.
## Acknowledgment
+
This package was first inspired by the standard, tested, and loved [MCML from Wang, Jacques and Zheng](https://omlc.org/software/mc/mcpubs/1995LWCMPBMcml.pdf) , itself based on [Prahl](https://omlc.org/~prahl/pubs/abs/prahl89.html) and completely documented, explained, dissected by [Jacques](https://omlc.org/software/mc/) and [Prahl](https://omlc.org/~prahl/pubs/abs/prahl89.html). The original idea of using Monte Carlo for tissue optics calculations was [first proposed by Wilson and Adam in 1983](https://doi.org/10.1118/1.595361). This would not be possible without the work of these pioneers.
diff --git a/assets/banner.png b/assets/banner.png
new file mode 100644
index 00000000..67fb5b7c
Binary files /dev/null and b/assets/banner.png differ
diff --git a/docs/README.assets/pytissue-demo-banenr.jpg b/docs/README.assets/pytissue-demo-banenr.jpg
deleted file mode 100644
index 8bf58cc7..00000000
Binary files a/docs/README.assets/pytissue-demo-banenr.jpg and /dev/null differ
 -
- -
- -
-