IDPEnsembleTools: An Open-Source Library for Analysis of Conformational Ensembles of Disordered Proteins
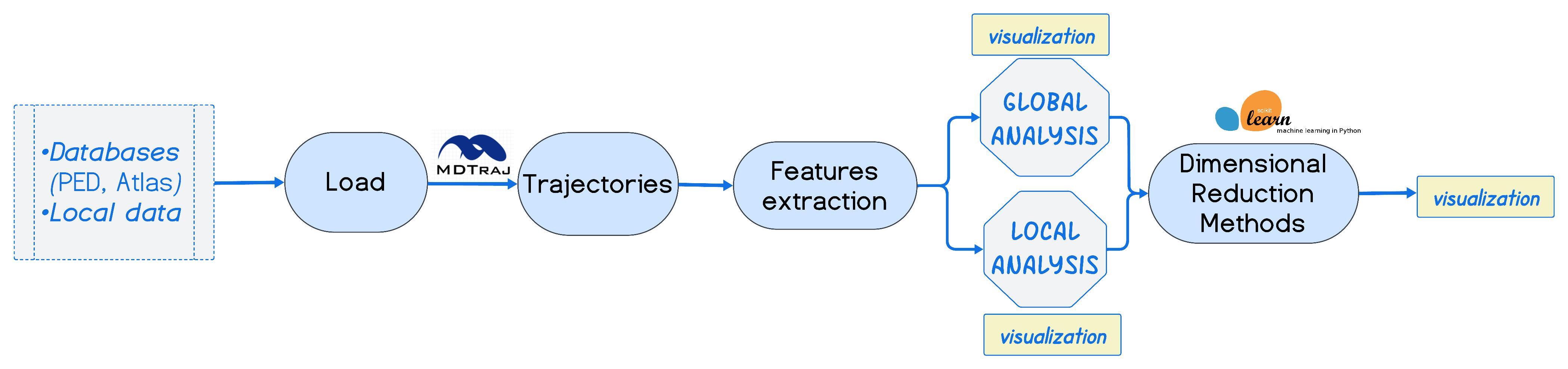
IDPEnsembleTools is a Python package designed to facilitate the loading, analysis, and comparison of multiple conformational ensembles of intrinsically disordered proteins (IDPs).
It supports various input formats such as .pdb, .xtc, and .dcd, and enables users to extract both global and local structural features, perform dimensionality reduction, and compute similarity scores between ensembles.
Full documentation is available at:
https://bioComputingUP.github.io/EnsembleTools
With IDPEnsembleTools, you can:
-
Extract global features of structural ensembles:
- Radius of gyration (Rg)
- Asphericity
- Prolateness
- End-to-end distance
-
Extract local features:
- Interatomic distances
- Phi–psi angles
- Alpha-helix content
-
Perform dimensionality reduction on ensemble features:
- PCA
- UMAP
- t-SNE
-
Compare structural ensembles using:
- Jensen-Shannon (JS) divergence
- Visualize similarity matrices
The notebooks/ directory contains a collection of Jupyter notebooks that demonstrate how to use the EnsembleTools package. These examples cover key functionalities such as ensemble comparison, dimensionality reduction (PCA, t-SNE, UMAP), feature extraction, and visualization customization. They serve both as tutorials and reproducible workflows for analyzing disordered protein ensembles.
| Notebook | Description | Link |
|---|---|---|
comparing_ensembles.ipynb |
Compare multiple conformational ensembles using selected metrics and visualizations. | View |
featurization.ipynb |
Generate numerical features from protein ensembles for downstream analysis. | View |
kpca_analysis.ipynb |
Perform Kernel PCA to capture non-linear variance in ensemble structures. | View |
loading_data.ipynb |
Load and preprocess ensemble data from various formats. | View |
pca_analysis.ipynb |
Principal Component Analysis (PCA) for dimensionality reduction and visualization. | View |
plot_customization.ipynb |
Customize plots for clarity and publication-quality visualizations. | View |
sh3_example.ipynb |
Case study: global and local analysis of the SH3 domain of the Drk protein. | View |
tsne_analysis.ipynb |
t-SNE embedding of ensemble features to explore local structure. | View |
umap_analysis.ipynb |
UMAP embedding of ensemble features and visualization. | View |
It is recommended to install idpet in a clean virtual environment to avoid conflicts with existing packages.
# Create and activate a new conda environment
conda create -n idpet-env python=3.9
conda activate idpet-env
# Install the package from PyPI
pip install idpet# Create a new virtual environment (Python 3.7+)
python -m venv idpet-env
# Activate the environment
# On Linux/macOS:
source idpet-env/bin/activate
# On Windows:
idpet-env\Scripts\activate
# Upgrade pip and install the package
pip install --upgrade pip
pip install idpet git clone https://github.com/BioComputingUP/EnsembleTools.git
cd idpet
pip install -e .This project is licensed under the MIT License.