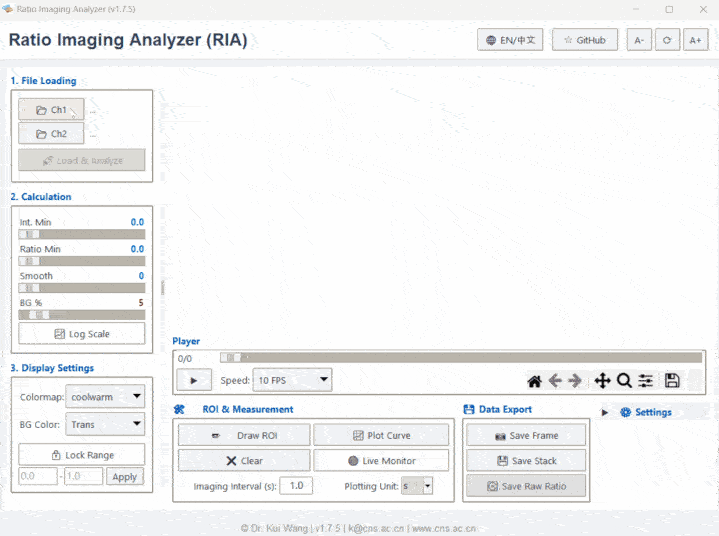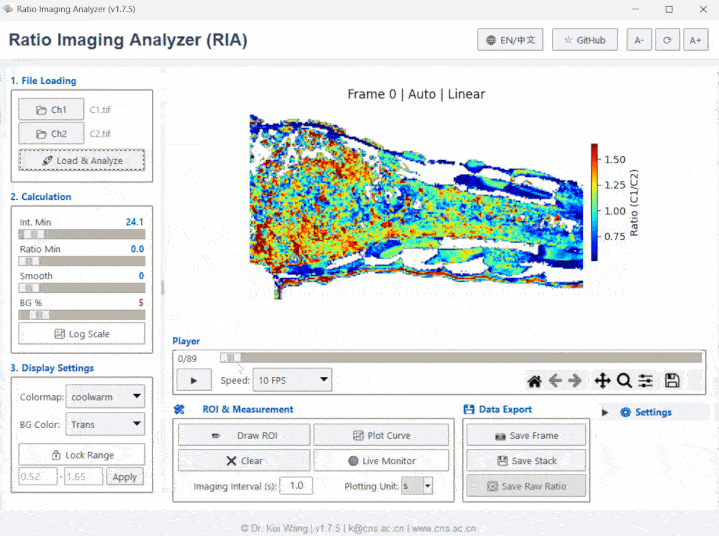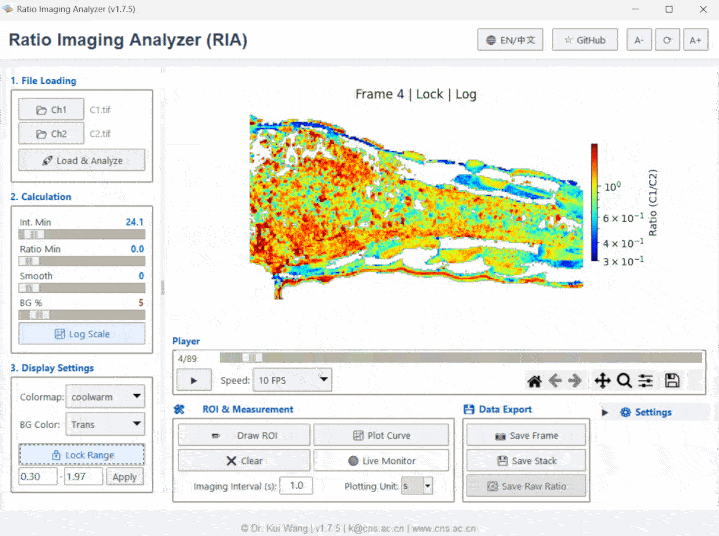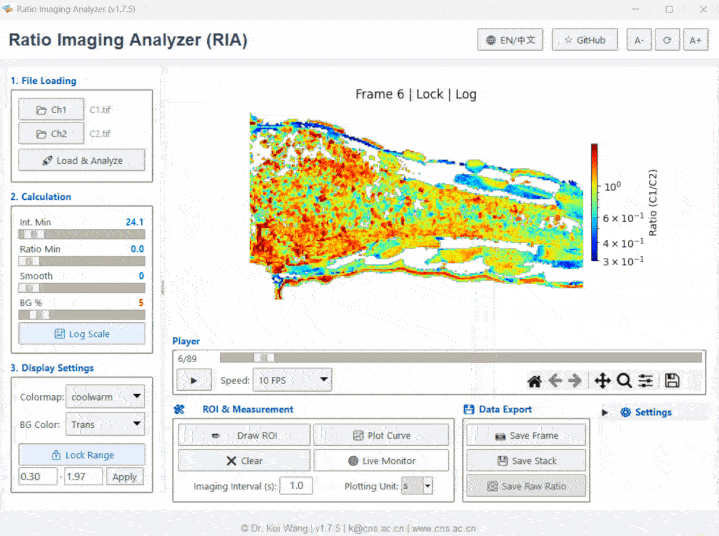
Meet RIA (or as we affectionately call her, "Li Ya / 莉丫").
Ratio Imaging Analyzer (RIA) is a specialized, standalone bio-image analysis software engineered for quantitative ratiometric microscopy. It provides a rigorous, coding-free workflow for processing dual-channel time-lapse data, serving as a powerful alternative to complex ImageJ/Fiji plugins or expensive commercial workstations (e.g., MetaMorph, NIS-Elements).
While originally designed for ratiometric imaging, RIA is fully optimized for Metabolic Imaging and Label-Free Intrinsic Fluorescence studies. It features built-in motion correction (image registration), background subtraction, and standardized data export, making it an essential tool for:
- FRET & Biosensors: Standard CFP/YFP, GFP/RFP, and novel energetic sensors.
- Metabolic Imaging: NAD(P)H/FAD redox ratios and metabolite sensors (ATP, Lactate).
- Label-Free / Structural Biology: Ratiometric Tryptophan (Trp) imaging for protein unfolding/conformational changes.
- Ion Sensing: Calcium (Fura-2, GCaMP), pH (BCECF), and Chloride imaging.
- Analysis Unchained: Stop queuing for the lab workstation. RIA is a standalone executable that runs on standard PCs.
- Reproducibility Ready: Science requires verification. RIA allows you to save your entire workspace (ROIs, thresholds, background settings) into a lightweight
.riaproject file. Send this file to collaborators or reviewers, allowing them to instantly reproduce your analysis and verify your results. - Math Done Right: Calculating ratios isn't just
A / B. Biological images have edges and noise. We implemented a normalized convolution algorithm that handlesNaN(Not a Number) values correctly. This means your data doesn't get eroded or corrupted at cell boundaries. - Zero Coding Needed: We know not everyone loves Python. RIA has a full GUI for background subtraction, thresholding, and dragging-and-dropping ROIs.
- Trust Your Data: We don't hide the numbers. You get the visual stacks, but you also get the raw float32 ratio data and time-series CSVs. You can take these straight to Prism, Origin, or Excel.
RIA is algorithmically agnostic to the specific fluorophores, making it compatible with any dual-emission or dual-excitation ratiometric modality:
- Intrinsic Tryptophan Fluorescence: Analyze protein conformational changes, unfolding, or binding events by calculating the ratio of emission shifts (e.g., 330nm / 350nm).
- Anisotropy-based Ratios: Process polarized emission channels to study molecular tumbling.
- Redox Ratio (Optical Redox Imaging): Quantify metabolic states (Glycolysis vs. OXPHOS) by computing the NADH / FAD autofluorescence ratio.
- Genetically Encoded Metabolite Sensors: Analyze modern biosensors for ATP (e.g., ATeam), Lactate, Pyruvate, or Glucose.
- Intermolecular / Intramolecular FRET: Robust processing for standard acceptor/donor emission stacks.
- Kinase Activity Reporters: Track phosphorylation events in real-time (e.g., AKAR, CKAR).
- Calcium Imaging: Ratiometric dyes (Fura-2, Indo-1) and sensors (Ratiometric-Pericam, GCaMP variants).
- pH & Chloride: BCECF, Clomeleon, and other environmental probes.
RatioImagingAnalyzer/
├── data/ # Sample TIFFs so you can try it out immediately
├── paper/ # JOSS submission files
├── src/ria_gui # The actual code
│ ├── main.py # Start here
│ ├── gui.py # The frontend logic
│ ├── processing.py # The math/algorithm heavy lifting
│ └── components.py # UI Widgets
├── tests/ # Automated tests to keep bugs away
└── requirements.txt # Dependencies
RIA is available in two editions to suit different needs: RIA Pro (for advanced features) and RIA Lite (for portability).
Recommended for: Researchers working with .oir, .nd2, .czi files.
The Pro version is the full-featured Python package. It includes comprehensive dependencies (aicsimageio, bioformats) to support reading professional microscopy formats directly.
⚠️ Prerequisite: Java (JDK) To read proprietary formats (like.oir,.nd2) via the Bio-Formats backend, you must have a Java Development Kit (JDK) installed on your system (OpenJDK 11 or later is recommended).
- Exclusive Feature: Direct support for Olympus .oir, Nikon .nd2, and Zeiss .czi files.
- Installation:
Once installed, launch it with:
pip install ria-gui
ria
Recommended for: Users who want instant access without installing Python.
The Lite version is a re-engineered, lightweight executable optimized for speed and portability. We have significantly reduced the file size and optimized the initialization process for instant startup.
- Best For: Standard TIFF workflows on any Windows PC.
- No Java Required: Since Lite handles standard TIFFs, no external Java environment is needed.
- Key Features: Zero configuration, ultra-fast cold start, minimal memory footprint.
- Download: Check the Releases page to download the latest
RIA_Lite_vX.X.exe.
For developers who want to contribute or modify the code:
-
Clone the repository:
git clone [https://github.com/Epivitae/RatioImagingAnalyzer.git](https://github.com/Epivitae/RatioImagingAnalyzer.git) cd RatioImagingAnalyzer -
Install dependencies: It is recommended to use a virtual environment.
pip install -r requirements.txt
-
Run the application:
python src/ria_gui/main.py
- Load Files:
- Supports both Single-Channel (Intensity) and Multi-Channel (Ratio) Tiff stacks.
- RIA Pro users can directly drag & drop
.oir/.nd2files (ensure Java is installed).
- Preprocessing:
- Motion Correction: Align shaky time-lapse data using the built-in ECC algorithm.
- Background: Set a global background subtraction (Percentile) or use a custom ROI.
- Visualization:
- Switch views between Ratio, Ch1, Ch2, or Aux channels using the toolbar.
- Analyze:
- Draw ROIs (Rectangle, Circle, Polygon).
- Click Plot Curve to see real-time intensity/ratio changes.
- Save & Export:
- Save Project: Save your session as a
.riafile to allow others to reproduce your work. - Export Data: Copy data to clipboard or save processed images as Tiff stacks.
- Save Project: Save your session as a
RIA v1.8.0 introduces a "Ghost Pilot" automated testing script powered by rich to ensure stability.
To run the visual E2E test demo:
python tests/auto_drive_rich.pyContributions are welcome! Please check the Issue Tracker or submit a Pull Request.
Distributed under the MIT License. See LICENSE for more information.
If you use RIA in your research, please cite:
Wang, K. (2025). Ratio Imaging Analyzer (RIA): A Lightweight, Standalone Python Tool for Portable Fluorescence Analysis (v1.8.3). Zenodo. https://doi.org/10.5281/zenodo.18107966
Or use the BibTeX entry:
@software{Wang_RIA_2025,
author = {Wang, Kui},
title = {{Ratio Imaging Analyzer (RIA): A Lightweight, Standalone Python Tool for Portable Fluorescence Analysis}},
month = dec,
year = {2025},
publisher = {Zenodo},
version = {v1.8.3},
doi = {10.5281/zenodo.18091693},
url = {[https://doi.org/10.5281/zenodo.18107966](https://doi.org/10.5281/zenodo.18091693)}
}