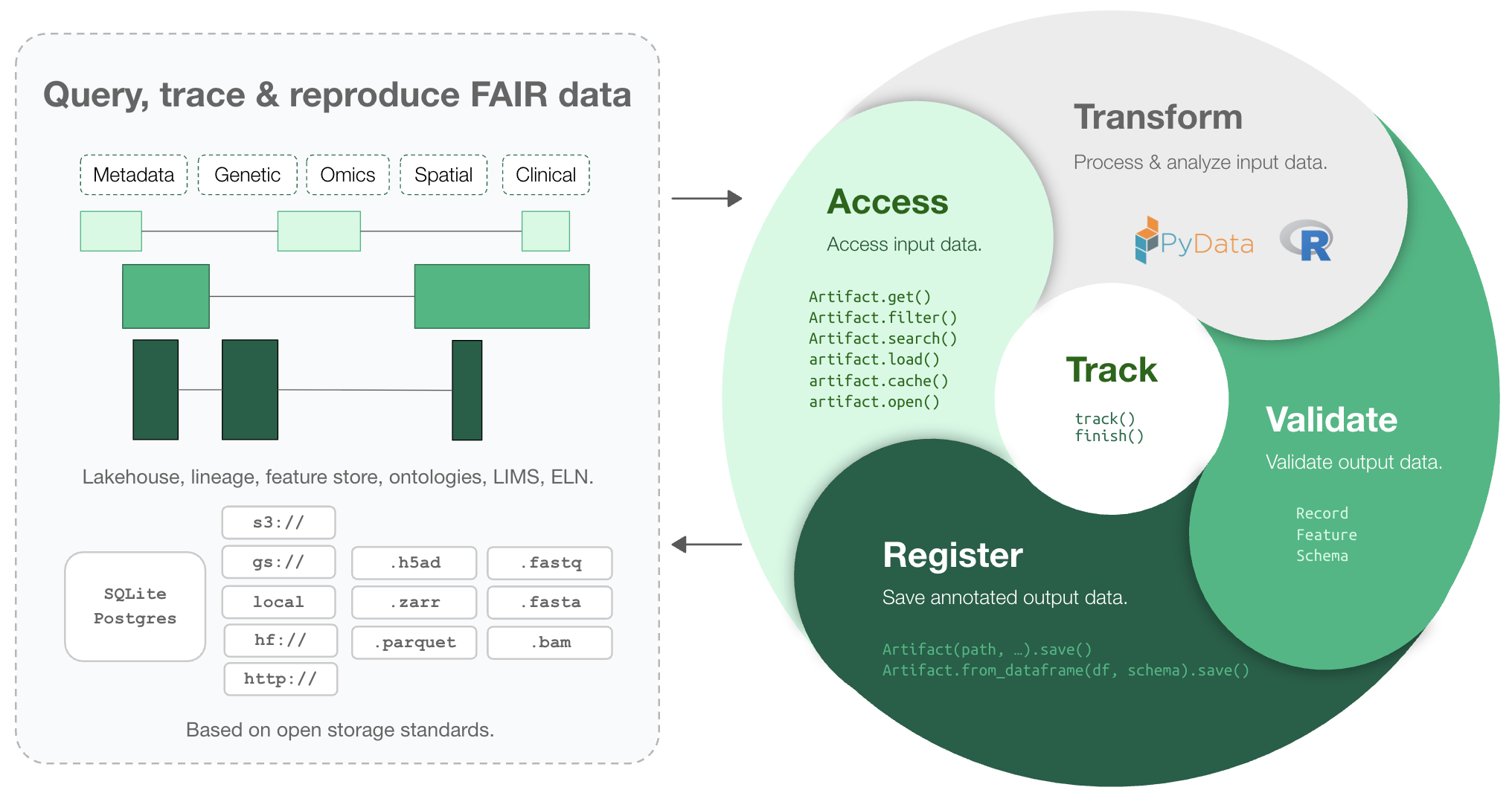
Makes your data queryable, traceable, reproducible, and FAIR. One API: lakehouse, lineage, feature store, ontologies, LIMS, ELN.
Why?
Reproducing analytical results or understanding how a dataset or model was created can be a pain. Training models on historical data, LIMS & ELN systems, orthogonal assays, or datasets from other teams is even harder. Even maintaining an overview of a project's datasets & analyses is more difficult than it should be.
Biological datasets are typically managed with versioned storage systems, GUI-focused platforms, structureless data lakes, rigid data warehouses (SQL, monolithic arrays), or tabular lakehouses.
LaminDB extends the lakehouse architecture to biological registries & datasets beyond tables (DataFrame, AnnData, .zarr, .tiledbsoma, …) with enough structure to enable queries and enough freedom to keep the pace of R&D high.
Moreover, it provides context through data lineage -- tracing data and code, scientists and models -- and abstractions for biological domain knowledge and experimental metadata.
Highlights:
- lineage → track inputs & outputs of notebooks, scripts, functions & pipelines with a single line of code
- lakehouse → manage, monitor & validate schemas; query across many datasets
- feature store → manage features & labels; leverage batch loading
- FAIR datasets → validate & annotate
DataFrame,AnnData,SpatialData,parquet,zarr, … - LIMS & ELN → manage experimental metadata, ontologies & markdown notes
- unified access → storage locations (local, S3, GCP, …), SQL databases (Postgres, SQLite) & ontologies
- reproducible → auto-version & timestamp execution reports, source code & environments
- zero lock-in & scalable → runs in your infrastructure; not a client for a rate-limited REST API
- integrations → vitessce, nextflow, redun, and more
- extendable → create custom plug-ins based on the Django ORM
If you want a GUI, you can connect your LaminDB instance to LaminHub and close the drylab-wetlab feedback loop: lamin.ai.
Who uses it?
Scientists & engineers in pharma, biotech, and academia, including:
- Pfizer – A global BigPharma company with headquarters in the US
- Ensocell Therapeutics – A BioTech with offices in Cambridge, UK, and California
- DZNE – The National Research Center for Neuro-Degenerative Diseases in Germany
- Helmholtz Munich – The National Research Center for Environmental Health in Germany
- scverse – An international non-profit consortium for open-source omics data tools
- The Global Immunological Swarm Learning Network – Research hospitals at U Bonn, Harvard, MIT, Stanford, ETH Zürich, Charite, Mount Sinai, and others
Copy summary.md into an LLM chat and let AI explain or read the docs.
Install the lamindb Python package:
pip install lamindbCreate a LaminDB instance:
lamin init --modules bionty --storage ./quickstart-data # or s3://my-bucket, gs://my-bucketOr if you have write access to an instance, connect to it:
lamin connect account/nameCreate a dataset while tracking source code, inputs, outputs, logs, and environment:
import lamindb as ln
ln.track() # track execution of source code as a run
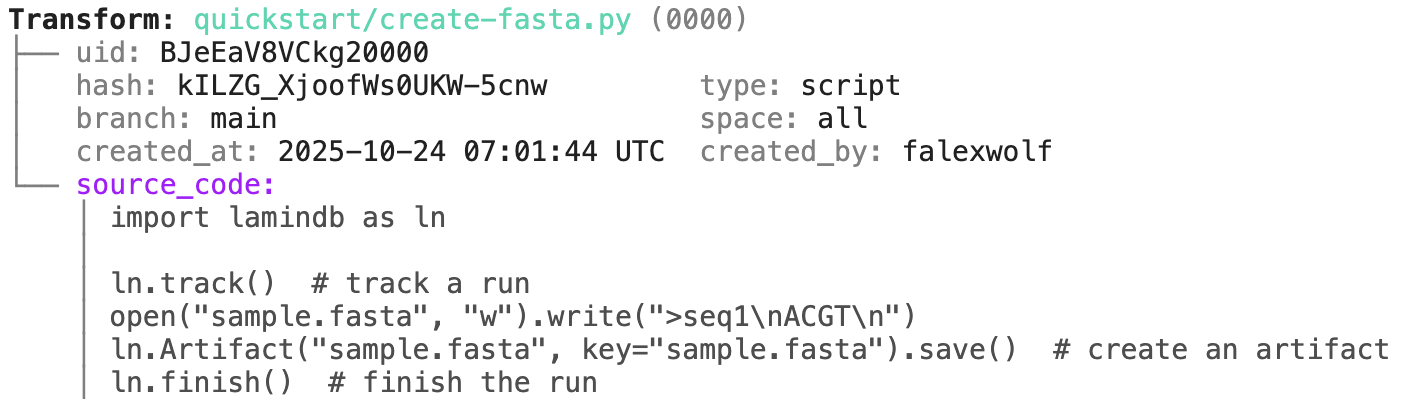
open("sample.fasta", "w").write(">seq1\nACGT\n") # create a dataset
ln.Artifact("sample.fasta", key="sample.fasta").save() # save dataset as an artifact
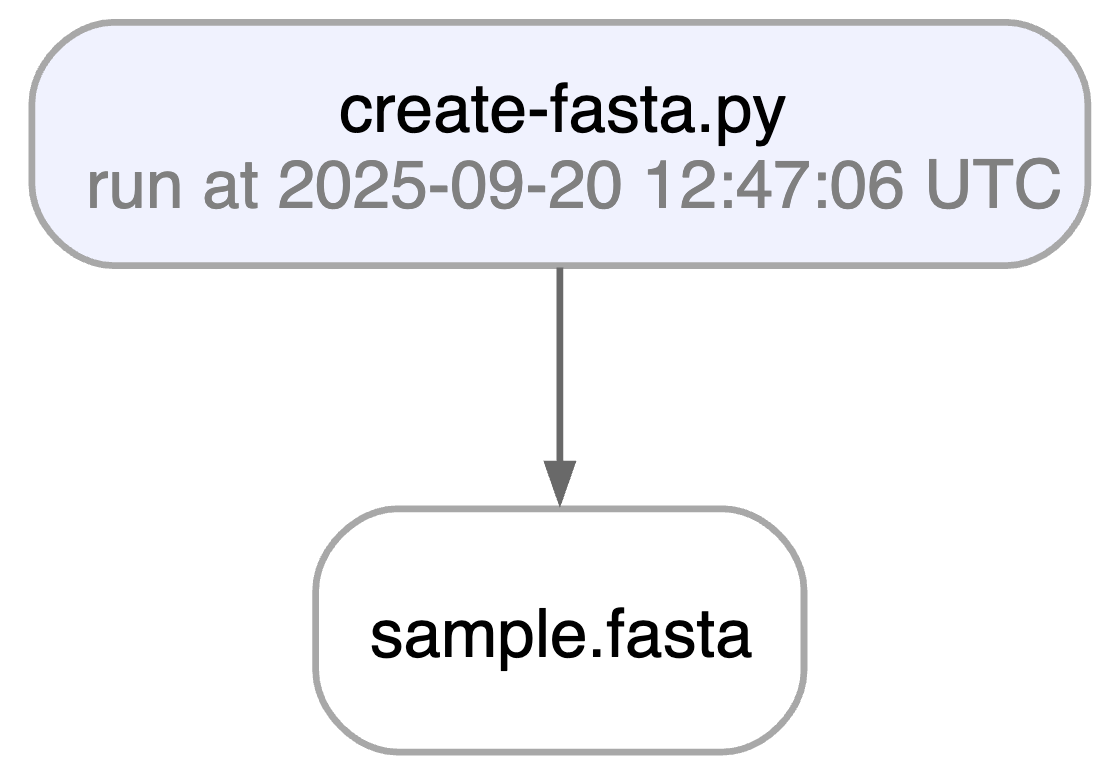
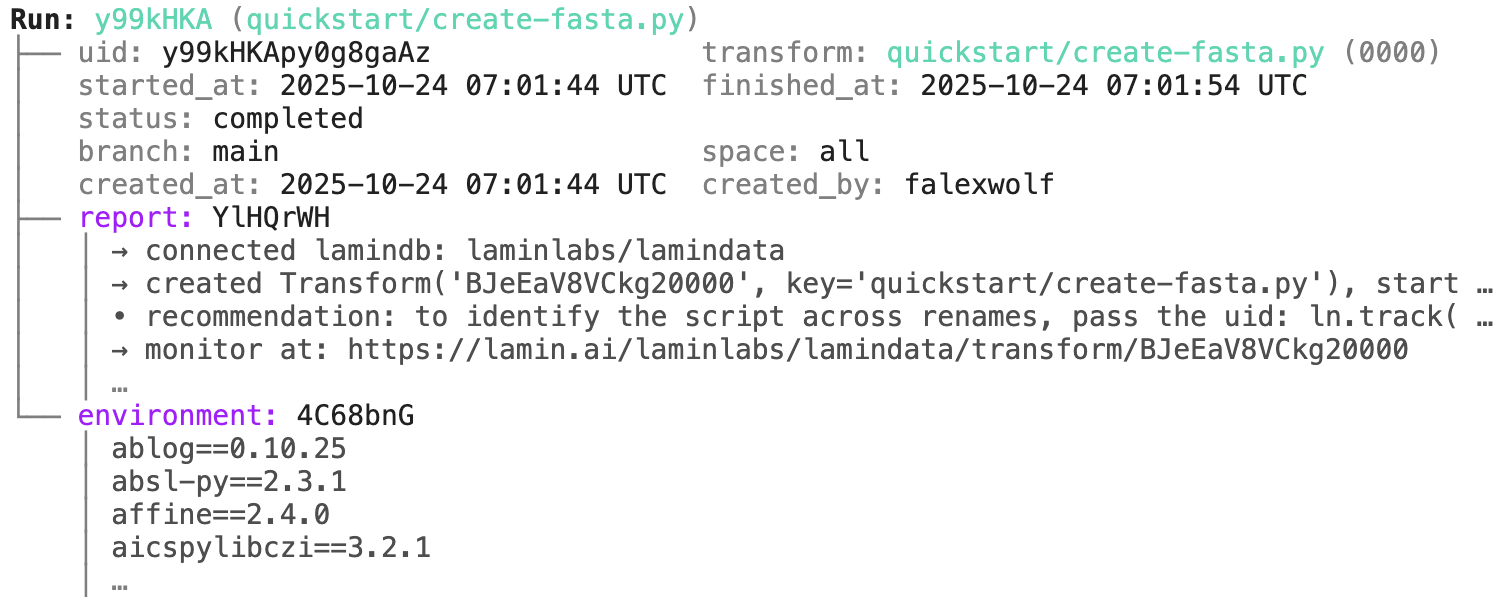
ln.finish() # mark the run as finishedRunning this snippet as a script (python create-fasta.py) produces the following data lineage.
artifact = ln.Artifact.get(key="sample.fasta") # get artifact by key
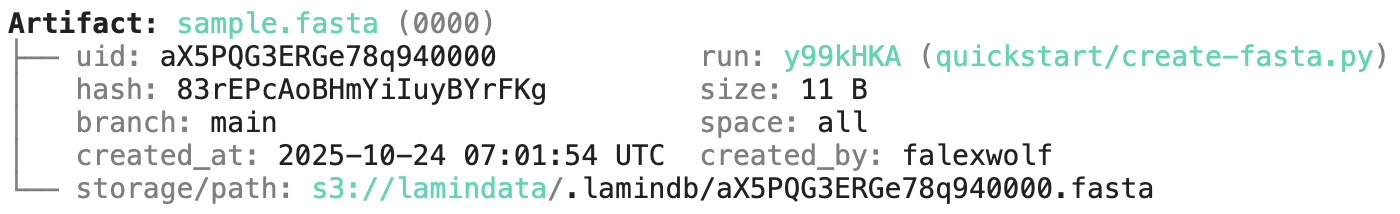
artifact.view_lineage()You'll know how that artifact was created and what it's used for. Basic metadata was captured in fields:
artifact.size # access the size
artifact.created_at # access the timestamp
# etc.
artifact.describe() # describe metadataHere is how to access the content of the artifact:
local_path = artifact.cache() # return a local path from a cache
object = artifact.load() # load object into memory
accessor = artifact.open() # return a streaming accessorAnd here is how to access its data lineage context:
run = artifact.run # get the run record
transform = run.transform # get the transform recordJust like artifacts, runs & transforms are SQLRecord objects and follow the same API, just with different fields.
run.describe() |
transform.describe() |
|---|---|
 |
 |
You can annotate datasets and samples with features. Let's define some:
from datetime import date
ln.Feature(name="gc_content", dtype=float).save()
ln.Feature(name="experiment_note", dtype=str).save()
ln.Feature(name="experiment_date", dtype=date).save()During annotation, feature names and data types are validated against these definitions:
artifact.features.add_values({
"gc_content": 0.55,
"experiment_note": "Looks great",
"experiment_date": "2025-10-24",
})Now that the data is annotated, you can query for it:
ln.Artifact.filter(experiment_date="2025-10-14").to_dataframe() # query all artifacts annotated with `experiment_date`You can also query by the metadata that lamindb automatically collects:
ln.Artifact.filter(run=run).to_dataframe() # query all artifacts created by a run
ln.Artifact.filter(transform=transform).to_dataframe() # query all artifacts created by a transform
ln.Artifact.filter(size__gt=1e6).to_dataframe() # query all artifacts bigger than 1MBIf you want to include more information into the resulting dataframe, pass include.
ln.Artifact.to_dataframe(include="features") # include the feature annotations
ln.Artifact.to_dataframe(include=["created_by__name", "storage__root"]) # include fields from related registriesYou can create records for the entities underlying your experiments: samples, perturbations, instruments, etc.. For example:
sample_type = ln.Record(name="Sample", is_type=True).save() # a sample type
ln.Record(name="P53mutant1", type=sample_type).save() # sample 1
ln.Record(name="P53mutant2", type=sample_type).save() # sample 2Define the corresponding features and annotate:
ln.Feature(name="design_sample", dtype=sample_type).save()
artifact.features.add_values({"design_sample": "P53mutant1"})You can query & search the Record registry in the same way as Artifact or Run.
ln.Record.search("p53").to_dataframe()You can also create relationships of entities and -- if you connect your LaminDB instance to LaminHub -- edit them like Excel sheets in a GUI.
If you change source code or datasets, LaminDB manages their versioning for you.
Assume you run a new version of our create-fasta.py script to create a new version of sample.fasta.
import lamindb as ln
ln.track()
open("sample.fasta", "w").write(">seq1\nTGCA\n") # a new sequence
ln.Artifact("sample.fasta", key="sample.fasta", features={"design_sample": "P53mutant1"}).save() # annotate with the new sample
ln.finish()If you now query by key, you'll get the latest version of this artifact.
artifact = ln.Artifact.get(key="sample.fasta") # get artifact by key
artifact.versions.to_dataframe() # see all versions of that artifactHere is how you ingest a DataFrame:
import pandas as pd
df = pd.DataFrame({
"sequence_str": ["ACGT", "TGCA"],
"gc_content": [0.55, 0.54],
"experiment_note": ["Looks great", "Ok"],
"experiment_date": ["2025-10-24", "2025-10-25"],
})
ln.Artifact.from_dataframe(df, key="my_datasets/sequences.parquet").save() # no validationTo validate & annotate the content of the dataframe, use a built-in schema:
ln.Feature(name="sequence_str", dtype=str).save() # define a remaining feature
artifact = ln.Artifact.from_dataframe(
df,
key="my_datasets/sequences.parquet",
schema="valid_features" # validate columns against features
)
artifact.describe()Now you know which schema the dataset satisfies. You can filter for datasets by schema and then launch distributed queries and batch loading.
To validate an AnnData with a built-in schema call:
import anndata as ad
import numpy as np
adata = ad.AnnData(
X=pd.DataFrame([[1]*10]*21).values,
obs=pd.DataFrame({'cell_type_by_model': ['T cell', 'B cell', 'NK cell'] * 7}),
var=pd.DataFrame(index=[f'ENSG{i:011d}' for i in range(10)])
)
artifact = ln.Artifact.from_anndata(
adata,
key="my_datasets/scrna.h5ad",
schema="ensembl_gene_ids_and_valid_features_in_obs"
)
artifact.describe()To validate a spatialdata or any other array-like dataset, you need to construct a Schema. You can do this by composing the schema of a complicated object from simple pandera/pydantic-like schemas: docs.lamin.ai/curate.
Plugin bionty gives you >20 of them as SQLRecord registries. This was used to validate the ENSG ids in the adata just before.
import bionty as bt
bt.CellType.import_source() # import the default ontology
bt.CellType.to_dataframe() # your extendable cell type ontology in a simple registryMost of the functionality that's available in Python is also available on the command line (and in R through LaminR). For instance, to upload a file or folder, run:
lamin save myfile.txt --key examples/myfile.txtLaminDB is not a workflow manager, but it integrates well with existing workflow managers and can subsitute them in some settings.
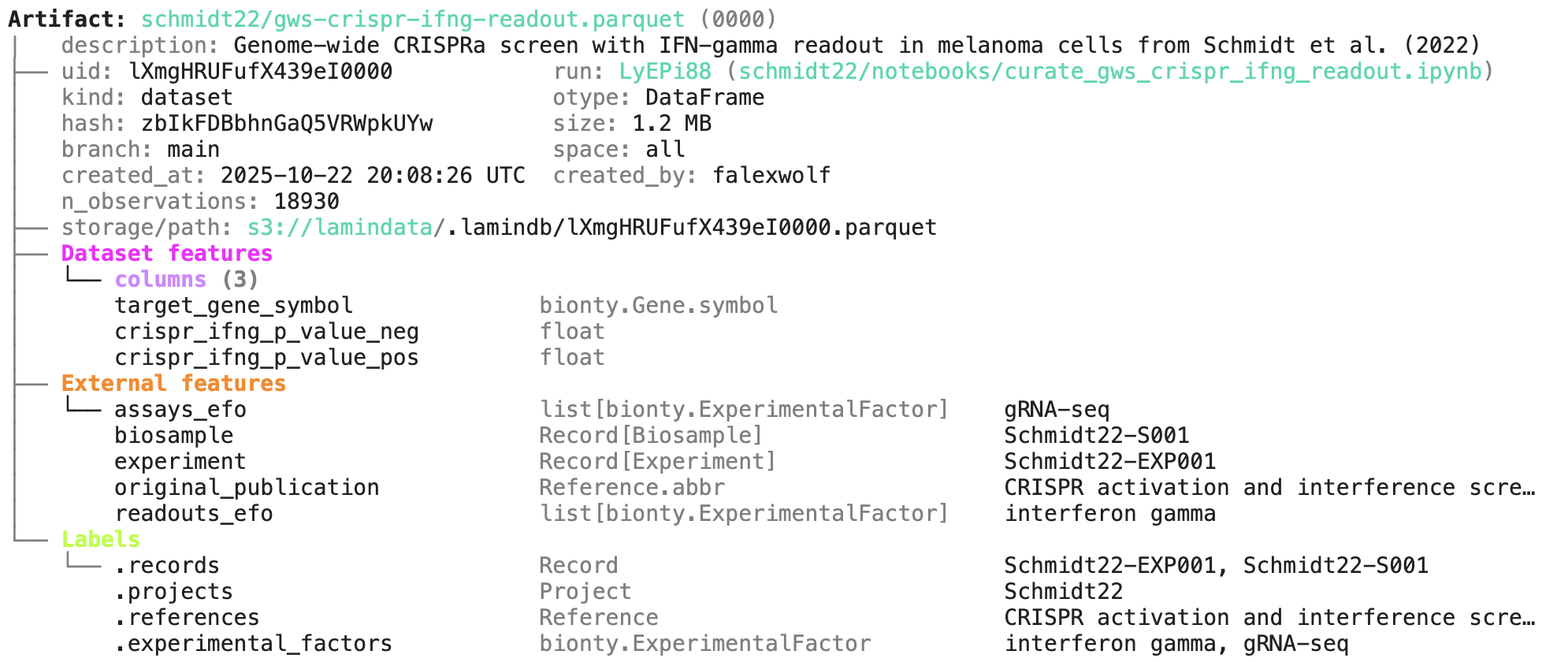
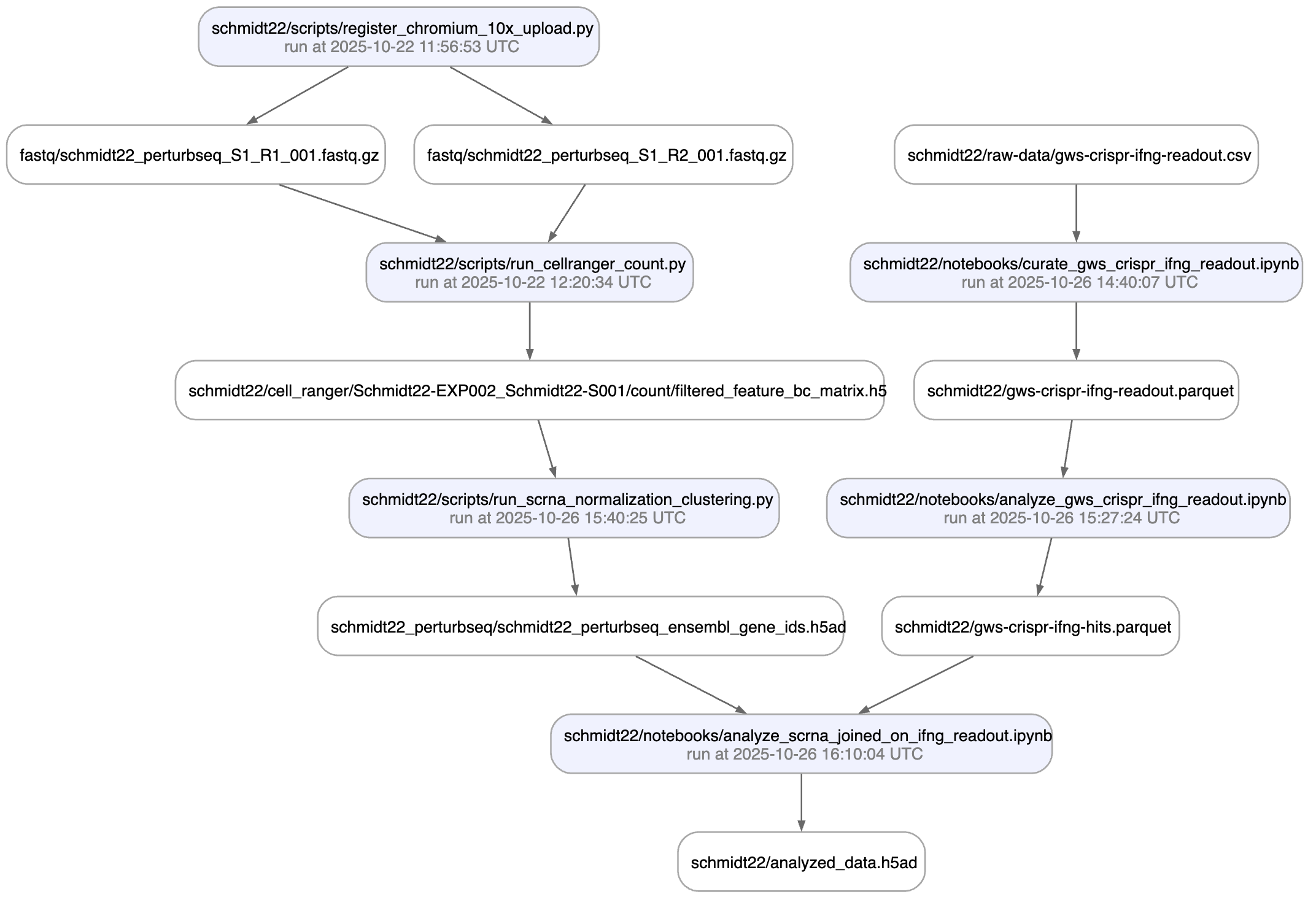
In github.com/laminlabs/schmidt22 we manage several workflows, scripts, and notebooks to re-construct the project of Schmidt el al. (2022). A phenotypic CRISPRa screening result is integrated with scRNA-seq data. Here is one of the input artifacts:
And here is the lineage of the final result:
You can explore it here.
If you'd like to integrate with Nextflow, Snakemake, or redun, see here: docs.lamin.ai/pipelines