Fast change point detection in Python
A Python library for detecting change points in time series and sequential data using PELT (Pruned Exact Linear Time) and SeGD (Sequential Gradient Descent) algorithms.
Documentation: https://zhangxiany-tamu.github.io/fastcpd_Python/
Now available on Test PyPI for testing! Try it with:
pip install --index-url https://test.pypi.org/simple/ --extra-index-url https://pypi.org/simple/ pyfastcpdFeatures:
- Parametric models: mean/variance, GLM (binomial/Poisson), linear/LASSO regression, ARMA/GARCH
- Nonparametric methods: rank-based and RBF kernel
- C++ implementation for core models, Python for specialized models
- Comprehensive evaluation metrics and dataset generators
- Optional Numba acceleration for GLM models
The package is available on Test PyPI as pyfastcpd:
# Install Armadillo first (required for building C++ extension)
# macOS:
brew install armadillo
# Ubuntu/Debian:
# sudo apt-get install libarmadillo-dev
# Then install pyfastcpd
pip install --index-url https://test.pypi.org/simple/ --extra-index-url https://pypi.org/simple/ pyfastcpdNote: Test PyPI currently only has source distribution (no pre-built wheels). You must install Armadillo before running pip install.
System Requirements for Building from Source:
- Python ≥ 3.8
- C++17 compiler
- Armadillo library (required for C++ compilation):
- macOS:
brew install armadillo - Ubuntu/Debian:
sudo apt-get install libarmadillo-dev
- macOS:
# Install Armadillo first (required!)
# macOS:
brew install armadillo
# Ubuntu/Debian:
# sudo apt-get install libarmadillo-dev
# Clone repository
git clone https://github.com/zhangxiany-tamu/fastcpd_Python.git
cd fastcpd_Python
# Install with editable mode
pip install -e .
# Optional extras for examples/benchmarks/time series notebooks
pip install -e .[dev,test,benchmark,timeseries]
# Optional: Install Numba for 7-14x GLM speedup
pip install numbaSupported Platforms: Linux, macOS (Windows support is experimental)
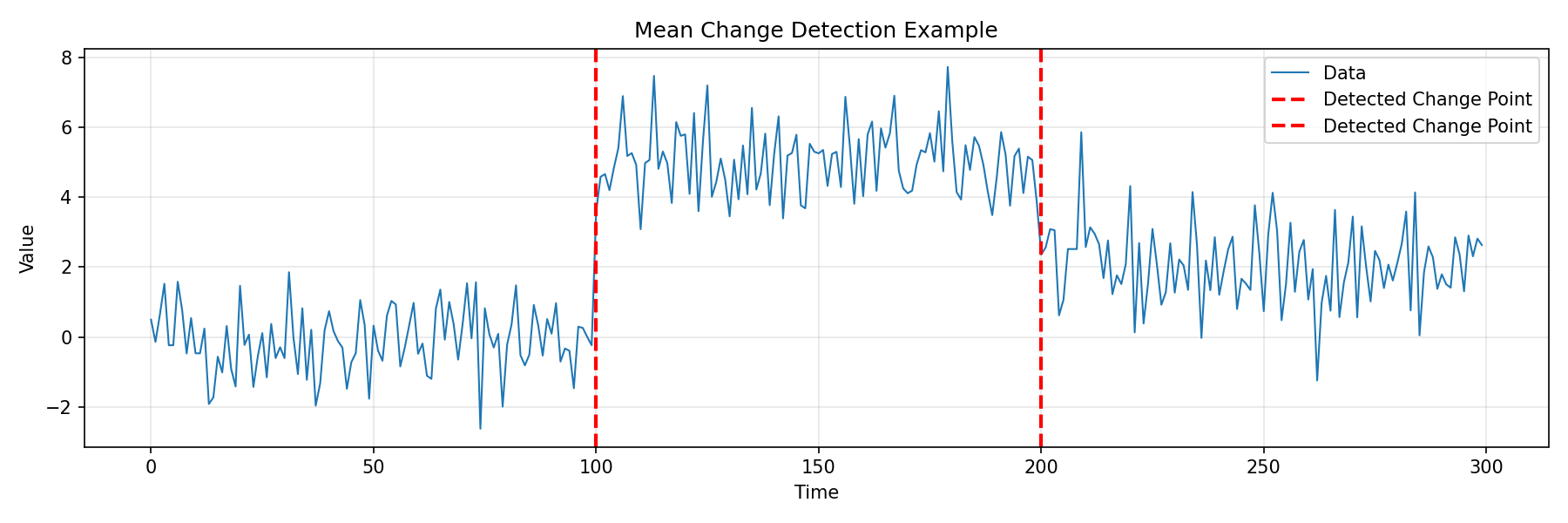
import numpy as np
import matplotlib.pyplot as plt
from fastcpd.segmentation import mean
# Generate data with mean changes
np.random.seed(42)
data = np.concatenate([
np.random.normal(0, 1, 100),
np.random.normal(5, 1, 100),
np.random.normal(2, 1, 100)
])
# Detect change points
result = mean(data, beta="MBIC")
print(f"Detected change points: {result.cp_set}")
# Output: [100, 200]
# Plot
plt.figure(figsize=(12, 4))
plt.plot(data, label='Data', linewidth=1)
for cp in result.cp_set:
plt.axvline(cp, color='red', linestyle='--', linewidth=2, label='Detected Change Point')
plt.xlabel('Time')
plt.ylabel('Value')
plt.title('Mean Change Detection Example')
plt.legend()
plt.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('mean_change_detection.png', dpi=150)
plt.show()import numpy as np
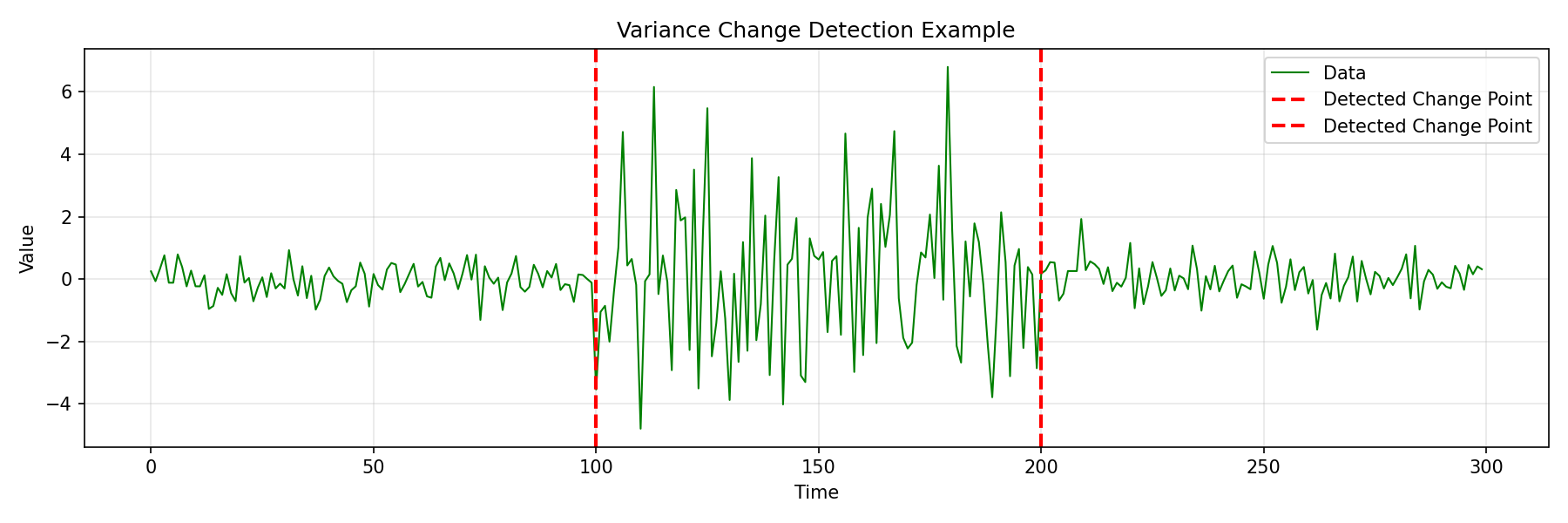
import matplotlib.pyplot as plt
from fastcpd.segmentation import variance
# Generate data with variance changes
np.random.seed(42)
data = np.concatenate([
np.random.normal(0, 0.5, 100),
np.random.normal(0, 2.5, 100),
np.random.normal(0, 0.5, 100)
])
# Detect change points
result = variance(data, beta="MBIC")
print(f"Detected change points: {result.cp_set}")
# Output: [100, 200]
# Plot
plt.figure(figsize=(12, 4))
plt.plot(data, label='Data', linewidth=1, color='green')
for cp in result.cp_set:
plt.axvline(cp, color='red', linestyle='--', linewidth=2, label='Detected Change Point')
plt.xlabel('Time')
plt.ylabel('Value')
plt.title('Variance Change Detection Example')
plt.legend()
plt.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('variance_change_detection.png', dpi=150)
plt.show()For combined mean and variance changes, use meanvariance(data, beta="MBIC")
Detect changes in autoregressive model parameters:
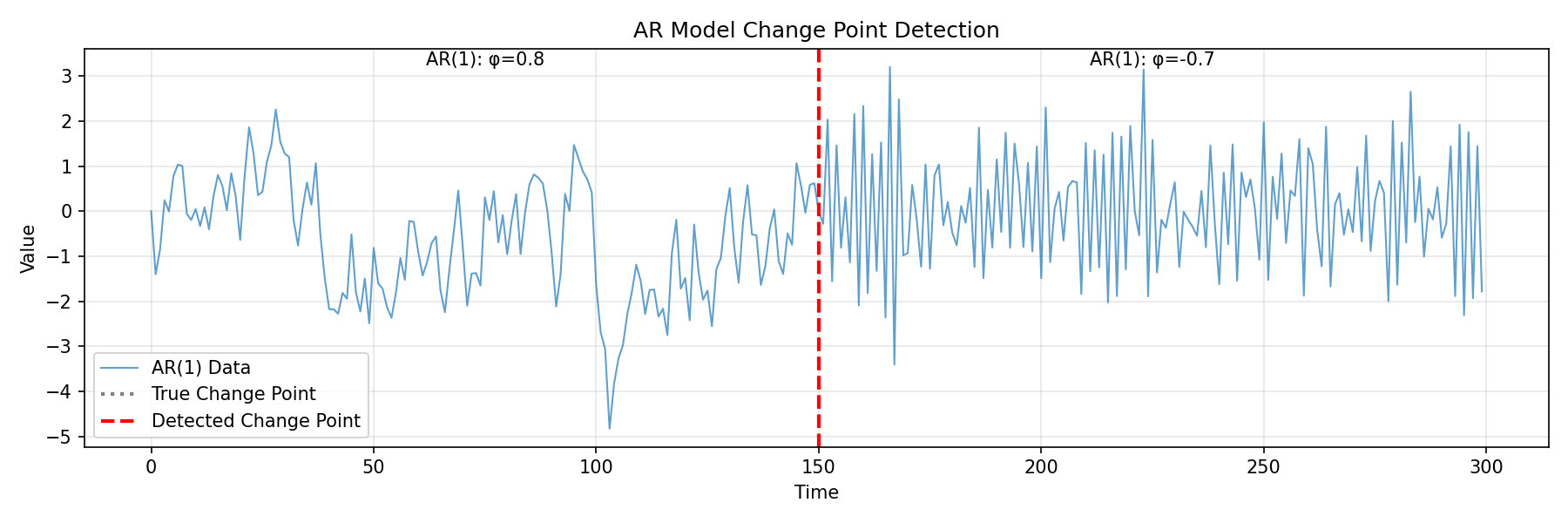
import numpy as np
import matplotlib.pyplot as plt
from fastcpd.segmentation import ar
# Generate AR(1) data with coefficient change
np.random.seed(100)
n = 300
# First segment: AR(1) with φ = 0.8
data1 = np.zeros(150)
for i in range(1, 150):
data1[i] = 0.8 * data1[i-1] + np.random.normal(0, 0.8)
# Second segment: AR(1) with φ = -0.7
data2 = np.zeros(150)
for i in range(1, 150):
data2[i] = -0.7 * data2[i-1] + np.random.normal(0, 0.8)
data = np.concatenate([data1, data2])
# Detect change points
result = ar(data, p=1, beta="MBIC")
print(f"Detected change points: {result.cp_set}")
# Output: [150]
# Plot
plt.figure(figsize=(12, 4))
plt.plot(data, label='AR(1) Data', linewidth=1, alpha=0.7)
plt.axvline(150, color='gray', linestyle=':', linewidth=2, label='True Change Point')
for cp in result.cp_set:
plt.axvline(cp, color='red', linestyle='--', linewidth=2, label='Detected Change Point')
plt.xlabel('Time')
plt.ylabel('Value')
plt.title('AR Model Change Point Detection')
plt.text(75, plt.ylim()[1]*0.9, 'AR(1): φ=0.8', ha='center')
plt.text(225, plt.ylim()[1]*0.9, 'AR(1): φ=-0.7', ha='center')
plt.legend()
plt.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('ar_change_detection.png', dpi=150)
plt.show()from fastcpd import fastcpd
# Logistic regression (binomial GLM)
# Data format: first column = response, remaining = predictors
data = np.column_stack([y, X])
result = fastcpd(data, family="binomial", beta="MBIC")
# Poisson regression
result = fastcpd(data, family="poisson", beta="MBIC")
# Linear regression
result = fastcpd(data, family="lm", beta="MBIC")
# LASSO (sparse regression)
result = fastcpd(data, family="lasso", beta="MBIC")# ARMA(p,q) - requires statsmodels
result = fastcpd(data, family='arma', order=[1, 1], beta='MBIC')
# GARCH(p,q) - requires arch package
result = fastcpd(data, family='garch', order=[1, 1], beta=2.0)from fastcpd.segmentation import rank, rbf
# Rank-based (distribution-free, monotonic-invariant)
result = rank(data, beta=50.0)
# RBF kernel (detects distributional changes via kernel methods)
result = rbf(data, beta=30.0)The vanilla_percentage parameter controls the PELT/SeGD hybrid:
# For GLM models only
result = fastcpd(data, family="binomial", vanilla_percentage=0.5)
# 0.0 = pure SeGD (fast), 1.0 = pure PELT (accurate), 'auto' = adaptive| Family | Description | Implementation |
|---|---|---|
mean |
Mean change detection | C++ (fast) |
variance |
Variance/covariance change | C++ (fast) |
meanvariance |
Combined mean & variance | C++ (fast) |
binomial |
Logistic regression | Python + Numba |
poisson |
Poisson regression | Python + Numba |
lm |
Linear regression | Python |
lasso |
L1-penalized regression | Python |
ar |
AR(p) autoregressive | Python |
var |
VAR(p) vector autoregressive | Python |
arma |
ARMA(p,q) time series | Python (statsmodels) |
garch |
GARCH(p,q) volatility | Python (arch) |
rank |
Rank-based (nonparametric) | Python |
rbf |
RBF kernel (nonparametric) | Python (RFF) |
Built-in Metrics:
- Precision, Recall, F1-Score
- Hausdorff distance, Covering metric
- Annotation error, One-to-one correspondence
Dataset Generators:
- Mean/variance shifts
- GLM coefficient changes
- Trend changes with multiple types
- Periodic/seasonal patterns
- Rich metadata for reproducibility
See examples/ and notebooks/ for demonstrations.
# Install system dependencies (macOS)
brew install armadillo
# Install system dependencies (Ubuntu/Debian)
sudo apt-get install libarmadillo-dev
# Clone and install
git clone https://github.com/zhangxiany-tamu/fastcpd_Python.git
cd fastcpd_Python
pip install -e .
# Optional: Install Numba for 7-14x GLM speedup
pip install numba- fastcpd R package - Original R implementation
- fastcpd Web App - Interactive Shiny interface
This project is licensed under the MIT License - see the LICENSE file for details.
- Documentation: https://zhangxiany-tamu.github.io/fastcpd_Python/
- Issues: GitHub Issues
- Email: zhangxiany@stat.tamu.edu